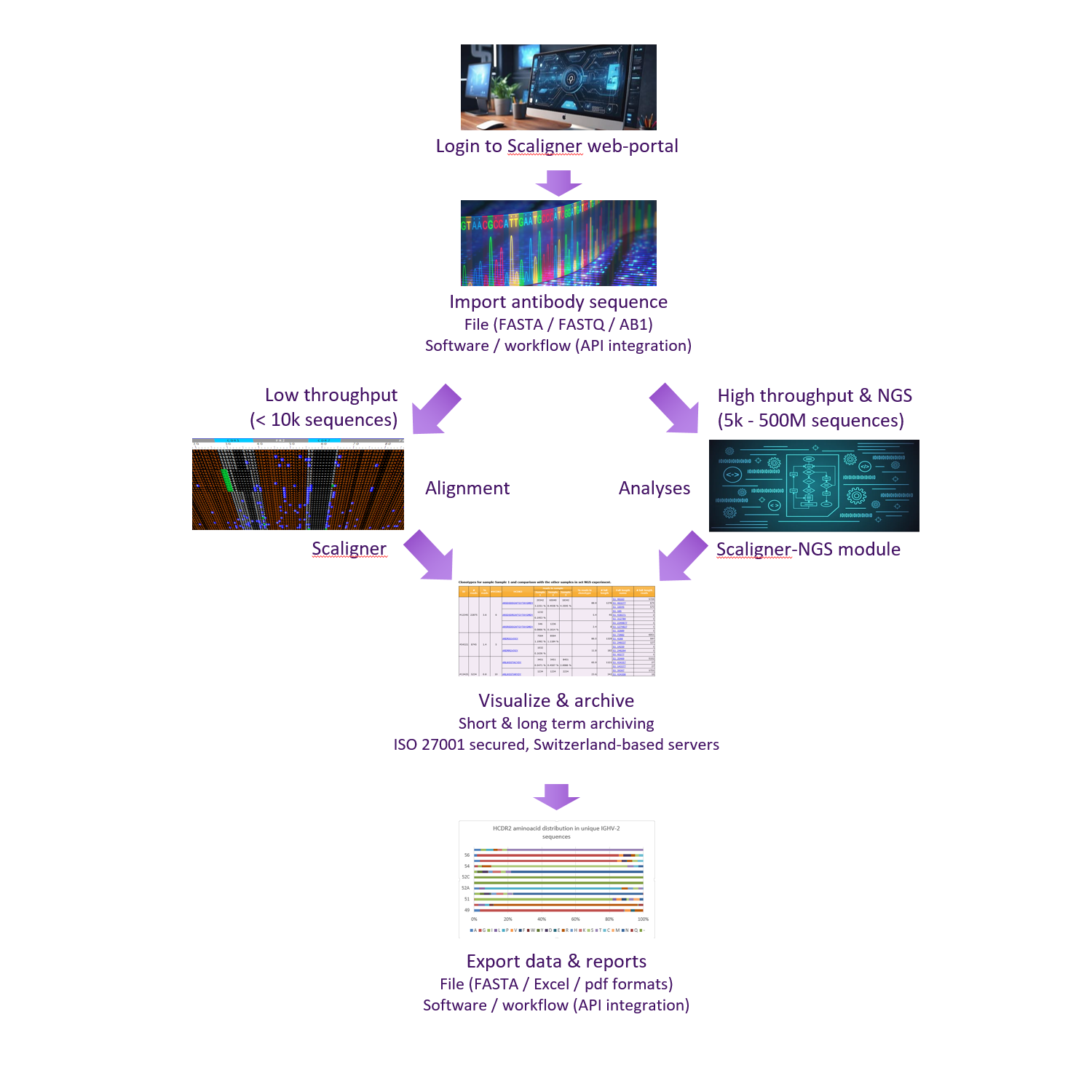
A seamless & adaptative workflow
Scaligner™ Cloud-based & On-Premises Platform






Germline identification
- Similarity scores
- Identification of closest germline
Identify liabilities
- Frameshifts, stop codons
- Post-translational modifications
- Mutation frequency
Clustering
- Group sequence by similarity
- Frequency analysis
Sequence alignment
- Sequence comparison
- Automatic CDR identification
Germline database
- Align against various species: human, rat, mice, rabbit, llama, etc
- Add custom germlines or references
Humaneness
- Predictability for similarity with mouse or human sequence
Scaligner-NGS: Next Generation Sequencing Module
The NGS analysis module integrates into your Scaligner™ software pipeline. Its powerful cloud computing platform will assist you to align millions of DNA sequences and extract actionable information from your sequence data at high speed.




